Unlocking the Future of Peptide-Based Precision Medicine
A 15-minute summary of our recent preprint, "PepTune: De Novo Generation of Therapeutic Peptides with Multi-Objective-Guided Discrete Diffusion"
Therapeutic peptides are on the rise, with over 33 FDA-approved therapeutic peptides introduced since 2000 and over 170 in clinical development. Unlike small molecules, peptides can bind to a diverse set of binding motifs present without requiring a stable binding pocket, making them optimal binders to structurally diverse protein targets. Their relatively large size and flexible backbone allow them to inhibit protein-protein interactions (PPIs) involved in disease processes that require binding to larger surface areas. Despite the recent success, the discovery of therapeutically viable peptide binders with high affinity for specific targets remains bottlenecked by traditional methods like phage screening, which requires screening trillions of random sequence permutations while still failing to cover the vast space of possible peptide sequences.
We sought to answer the question — how can we accelerate the search for peptides with necessary therapeutic properties? It is clear that generative AI is the perfect tool to narrow down the search space, so let’s dive deep into how we achieve this with PepTune, our multi-objective-guided discrete diffusion model for de novo generation of peptide SMILES.
If you are curious: “Pep” is for peptide and “Tune” is for tuning the properties of peptides with multi-objective guidance. And it sounds like Neptune.
In this post, I provide a brief background on discrete diffusion generative models. I summarize the key challenges in multi-objective peptide discovery and describe how we tackle these challenges with PepTune.
If you’re interested in learning more of the theory of the model, you can watch my talk at the Learning on Graphs and Geometry reading group, where I discuss our framework in depth.

Primer on Generative Diffusion Models
The fundamental idea behind generative deep learning models is simple: (1) train a parameterized model to learn a data distribution p(x), and (2) sample new data points from the learned distribution.
The key to understanding generative models lies in understanding Bayesian inference. Suppose we want to estimate the probability of a “clean” sequence (x) given a noisy sequence (z).
Intuitively, the probability of our clean sequence given the noisy sequence p(x|z) should be dependent on our prior knowledge of x and on the likelihood of obtaining the noisy sequence from some clean sequence.
Imagine we are guessing the contents of a clean image (represented by x) from a blurry image (represented by z). Intuitively, we are guessing based on some prior knowledge of what the image should be (e.g. “the image is of an animal”) as well as prior understanding of how clean images are converted to blurry images (e.g. “sharp edges become gradients when blurred”).
This idea leads to the following equation for p(x|z) which is known as Bayes’ theorem:
Let’s break down the terms in this equation:
p(x|z) is the conditional probability of a clean hypothesis z given the noisy features x. This is called the posterior probability and will determine how we convert noisy sequences into an output sequence.
p(z|x) is the likelihood of obtaining the noisy features from some clean hypothesis. In our example, this is our prior understanding of how clean images are converted to blurry images.
p(x) is the prior probability of the clean hypothesis x from a prior distribution. This is our prior knowledge of x or our prior knowledge of what the image should be.
p(z) is the distribution of the noisy features. This will often just be a Gaussian with unit-mean and zero-variance.
Bayes’ theorem tells us that the probability of obtaining a clean hypothesis from a set of noisy features is proportional to the prior probability of our clean hypothesis and the likelihood of obtaining the noisy features from some clean hypothesis. We can consider the distribution of noisy features in the denominator as a constant since it does not depend on the distribution of the clean features.
Diffusion models aim to iteratively reconstruct the clean sequence x from the noisy sequence z through a series of denoising steps.
Each denoising step is determined by a marginal reverse transition distribution, which we can define using Bayes’ rule.
Since we can’t simply draw noisy samples from thin air, we need to condition the distributions on some initial clean sequence from the training dataset x_0. The forward diffusion process adds noise to x_0 deterministically, which means we have the following closed-form equation for the ground-truth reverse transition distribution.
However, to generate de novo sequences, we don’t have access to a clean sequence x_0. Therefore, we train a parameterized deep learning model to predict the reverse transition distribution from only a noisy sequence z_t.
To train a model to accurately approximate the true reverse distribution, we must define a loss function such that by minimizing the loss, we minimize the discrepancy between the distribution of sequences generated from the model and the true data distribution. First, using our parameterized model, we define the Evidence Lower Bound (ELBO) of the log probability of generating a true sequence x_0.
By maximizing this lower bound, we are effectively maximizing how well the model is able to generate true samples x_0 from its learned distribution. Now, we can define our loss function as the negative ELBO (NELBO) such that by minimizing the loss function with gradient descent, we maximize the accuracy of the model.
As we derive in the paper, the NELBO can be expanded into the following form.
In short, a diffusion model learns to convert indistinguishable noise into a target “object” like a protein structure, a peptide sequence, or a picture of a cat (as shown below).

PepTune: Multi-Objective-Guided Masked Discrete Diffusion for Peptide SMILES
In summary, PepTune leverages Monte-Carlo Tree Search (MCTS) to guide a generative masked discrete diffusion model which iteratively refines a set of Pareto non-dominated sequences optimized across a set of therapeutic properties, including binding affinity, cell membrane permeability, solubility, non-fouling, and non-hemolysis.
Here, I will be providing a high-level overview of our model, but you can find all the theoretical and experimental details in our paper.
State-Dependent Masked Discrete Diffusion Model
Our first goal was to train a generative model that could generate valid peptide SMILES sequences. We achieved this by training a state-dependent masked discrete diffusion model on 11 million peptide SMILES.

At a high level, masked discrete diffusion models learn to accurately reconstruct “clean” sequences in the training set from the corrupted sequence with random [MASK] tokens with iterative unmasking steps.
Suppose you have a partially completed puzzle of a cat (since I’m a cat person). Given the pieces that are already in place and your prior understanding of what cats look like, you intuitively have an idea of what the missing pieces should look like. Therefore, you can easily discriminate between the leftover pieces that are likely and unlikely to fit into the remaining spots.
Similarly, a masked discrete diffusion model takes a partially unmasked sequence and given its previous “knowledge” of what peptide sequences should look like from the training data, predicts the likelihood of each token occurring at the [MASK] positions in the sequence. This allows us to generate peptide sequences completely from scratch by feeding a fully masked sequence into the model and iteratively unmasking the sequence by sampling from the predicted categorical distributions.
Read our paper for a deeper look into the theory behind our specific masked discrete diffusion formulation.
Multi-Objective-Guidance with Monte Carlo Tree Search
Given the range of properties required for a peptide to be a viable drug, our second goal was to achieve multi-objective guidance on a set of therapeutic properties. Since the multi-objective optimization problem must consider the infinite possible trade-offs between properties, we leverage the idea of Pareto non-dominance to search for peptide sequences that are maximally optimized across all objectives.
A Pareto non-dominated sequence is defined as a peptide sequence where none of the existing generated sequences are strictly better than it in at least one objective while being no worse in all remaining objectives. Intuitively, this means that we cannot improve a property of a non-dominated sequence without sacrificing another property. We define this idea formally in our paper.
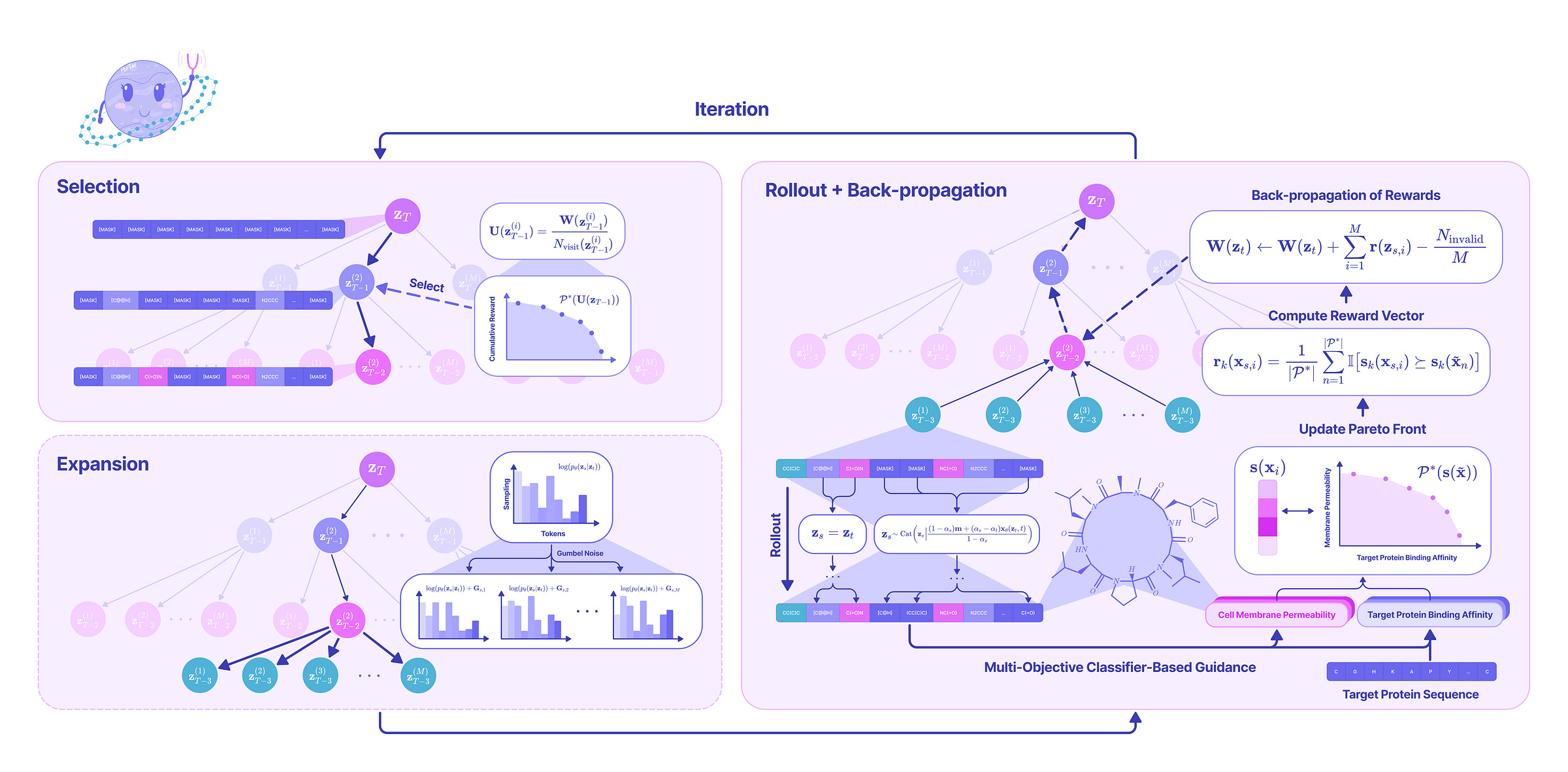
To find the Pareto non-dominated sequences, we leverage a novel classifier-based guidance strategy based on Monte Carlo Tree Search (MTCS) that iteratively executes the following steps:
Selection. From a fully masked sequence defining the root node of the MCTS tree, we traverse a path through the MCTS tree defined as a series of reverse unmasking steps that have been previously traversed based on the optimality of the sequences that were previously generated from that path.
Expansion. Upon reaching a terminal leaf node that has yet to be further unmasked, we perform batched unmasking where the sequence is unmasked in several distinct ways by sampling from the predicted reverse transition distribution from our trained masked discrete diffusion model.
Rollout. For each of the newly unmasked sequences in the batch, we greedily unmask the remaining masked tokens to get a set of fully unmasked sequences. Then, we evaluate the Pareto optimality of each sequence based on a vector of property scores generated from our pre-trained classifier models for binding affinity, membrane permeability, solubility, hemolysis, and non-fouling. All Pareto-optimal sequences generated in this step are stored in a global Pareto-optimal set that is returned at the end of the algorithm.
Backpropagation. The sum of the reward vectors generated from each child node in the batch is backpropagated to all predecessor nodes in the MCTS tree to bias the search towards unmasking paths that produce Pareto-optimal sequences.
At the end of a specified number of iterations, we return a full set of Pareto non-dominated sequences. From experiments with various protein targets and even multiple protein targets, we show that our strategy effectively optimizes scores across multiple properties simultaneously, generating strong peptide binders that meet the criteria for a viable drug.

Now that we have a general idea of the model architecture, let’s explore the gaps present in computational peptide discovery and how we fill these gaps with PepTune.
Gaps in Multi-Objective Peptide Discovery
Computational peptide discovery has recently taken off, with the development of structure-based models like RFpeptides and sequence-based models like PepPrCLIP and PepMLM. Despite this progress, we identified several limitations of existing models, which we address with PepTune.
Representing Non-Canonical and Cyclic Modifications
Existing generative peptide models that represent peptides as sequences of canonical amino acids fail to accommodate the representation of non-natural amino acids (nAAs) and cyclic peptides. The only relevant model we found, HELM-GPT, uses reinforcement learning to generate de novo peptide HELM sequences for KRAS binding affinity and membrane permeability. However, HELM notation encodes monomers as tokens, failing to fully capture the context of specific modifications and their structural features. Furthermore, the limited peptide data in HELM notation prevents the model from comprehensively learning the features of nAAs, which could bias generation towards natural AAs.
Successful drugs including selespressin and GLP-1 analogs contain a non-natural amino acids with longer half-life and stronger binding affinity to their target than their natural counterparts.
PepTune is the first generative model for peptide Simplified Molecular Input Line Entry System (SMILES) sequences which encode a higher level of granularity and can capture three-dimensional chemical structures, including non-canonical and cyclic modifications. To evaluate the validity of our generated peptides, we develop a platform that can parse a SMILES sequence, determine if it translates to a valid peptide containing peptide bonds, translate the SMILES string into an amino acid sequence from a library of over 200 ncAAs from SwissSidechain, and generate a two-dimensional visualization of the peptide. Try it out yourself on HuggingFace.
Granularity of SMILES Representations.
The precision of SMILES representations comes with its drawbacks. The vast majority of SMILES strings are either invalid molecules or do not translate into synthesizable peptides with peptide bonds, making generating valid peptide SMILES a significant challenge. To overcome this, PepTune leverages two core techniques: tokenization and state-dependent masking.
Tokenization merges single SMILES characters like ‘C’ (carbon atom) or ‘=’ (double bond) into common motifs across peptide SMILES like ‘NC(=O)’ (peptide bond). With a vocabulary generated specifically from a dataset of peptide SMILES, we can encourage the generation of common motifs in peptides while maintaining granularity through single-atom tokens.
State-dependent masking is a technique that varies the probability of transitioning into a [MASK] token during the forward diffusion process depending on the ‘state’ or token. By decreasing the masking rate of the backbone peptide bond tokens, the model learns to unmask the fundamental peptide-bond structure during generation before filling in the side-chain tokens. We show both theoretically and in practice that our strategy effectively increases the validity of the generated SMILES.
Multi-Objective Guidance in the Discrete State Space
Guided diffusion aims to guide the unmasking diffusion process towards favorable properties — like binding affinity or solubility in the context of therapeutic peptides. Guidance strategies can be broadly divided into two categories: classifier-free and classifier-based guidance.
Classifier-free guidance relies on high-quality, labeled datasets for specific properties, which fail in tasks where data is scarce like peptide sequences. Not to mention the need for data conditioned on multiple properties for multi-objective generation.
Classifier-based guidance that relies on smooth gradient updates based on a classifier score struggles in discrete state spaces where properties are evaluated on discrete sequences where the gradient is undefined.
The beauty of our Monte Carlo Tree Search (MTCS)-based strategy is that it requires no gradient estimation and performs optimization using defined rewards that are backpropagated to bias the model towards optimal unmasking strategies. By maintaining a global set of Pareto-optimal sequences and evaluating the optimality of a sequence on each objective separately with score vectors, we achieve multi-objective guidance without the risk of missing out on sequences that are optimized on some of the scores but not others. PepTune also does not require any additional training of the generative model when adding new objectives, providing a modular and scalable approach for multi-objective guidance.
Lack of High-Quality Property Classifiers for Peptide SMILES
How do we actually evaluate whether a sequence is optimal without testing it in vitro? Despite the abundance of deep learning models developed for predicting the properties of protein sequences and small molecule drugs, there was a significant gap in high-quality classifiers specifically trained on peptide SMILES sequences. So we took it upon ourselves to train classifiers for key peptide therapeutic properties including binding affinity, membrane permeability, solubility, non-hemolysis, and non-fouling.
For our binding affinity regressor, we took feature-rich pre-trained language model embeddings of the peptide and protein sequence combined with cross-attention layers to generate a binding affinity score based on peptide SMILES data from the PepLand database. For the remaining classifiers, we combined the feature-rich peptide embeddings and trained boosted tree models for logistic regression.

Despite achieving high accuracy on both the training and validation data, the limited high-quality labeled data for peptide SMILES remains a bottleneck to full generalizability.
What’s Next?
As we show in the paper, we have validated binders to diverse therapeutic targets including transferrin receptor (TfR) for blood-brain barrier targeting, GLP-1R for diabetes, glutamate-aspartate transporter (GLAST) for targeting astrocytes involved in neurological disorders, glial fibrillary acidic protein (GFAP) for Alexander disease, and anti-Müllerian hormone type-2 receptor (AMHR2) for polycystic ovarian syndrome.
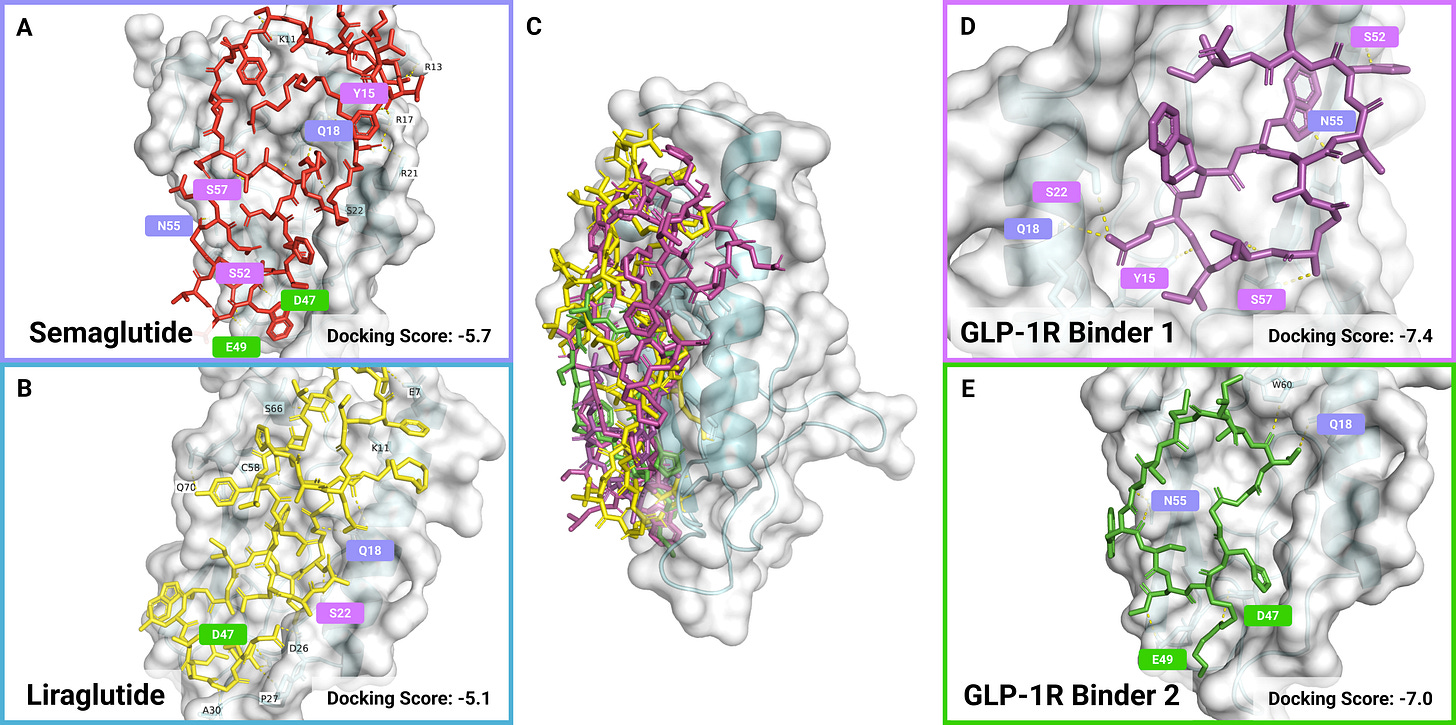
We also show that PepTune is capable of generating peptide binders that bind to multiple targets, which could have significant therapeutic applications. For instance, we generate binders with high affinity to both TfR and GLAST which could be used in intravenous drug delivery systems to enable delivery across the blood-brain barrier and specifically to astrocytes involved in several neurological diseases including Alzheimer’s disease and Parkinson’s disease. Alternatively, dual-targeting peptides can be used to degrade disease-causing proteins by binding to them and recruiting E3 ligases that tag the target protein with ubiquitin which induces proteasome degradation.
Given its unique generative capabilities, PepTune has endless therapeutic applications. First, PepTune requires no protein structure information, only a protein sequence, meaning it can generate binders for virtually any known protein target. Second, PepTune can condition on any set of therapeutic properties, ensuring that the candidates are good binders while meeting requirements for therapeutic viability. Finally, PepTune’s classifier-based guidance and generation occurs directly in the sequence space, ensuring that nothing is lost in translation either from structure to sequence or from continuous to discrete spaces.
Although our current peptides have only been validated with in silico docking, we are in the process of synthesizing our PepTune-generated peptides and conducting assays to validate target binding, solubility, and off-target binding (non-fouling). So stay tuned for updates to our preprint with new in vitro validation.
We believe PepTune is a first step towards a new era of computationally designed peptide-based precision medicine beyond just peptide drugs — from designing peptide guides for target protein degradation to ligands for targeted nanoparticle-based drug-delivery systems.
Citation
If you find this article helpful for your publications, please consider citing our paper:
@article{tang2024peptune,
title={PepTune: De Novo Generation of Therapeutic Peptides with Multi-Objective-Guided Discrete Diffusion},
author={Tang, Sophia and Zhang, Yinuo and Chatterjee, Pranam},
journal={42nd International Conference on Machine Learning},
year={2024}
}Thank you for reading! Special thanks to my co-authors and the Chatterjee Lab behind this work. Feel free to connect with me on LinkedIn to stay updated on new research or visit my website to learn more. Also, make sure to check out the full preprint on arXiv:
Alchemy Bio is a blog where I share transformational ideas in computational biology, synbio, and biotech. If you don’t want to miss upcoming blogs, you should consider subscribing for free to have them delivered to your inbox:


